| Description | Link to file | Date |
| Inputs |
| Submitted sequence(s) | NP_510002_1.a2m | 2010 Mar 11 20:35:37 |
| README file | README | 2010 Mar 11 20:35:55 |
| Quick blastp of non-redundant PDB | NP_510002_1.pdb_blast.txt | 2010 Mar 11 20:35:58 |
| Multiple alignment |
| SAM_t06 multiple alignment in a2m format | NP_510002_1.t06.a2m.gz | 2010 Mar 11 21:09:52 |
| SAM_t06 multiple alignment in pretty html format | NP_510002_1.t06.pa.html | 2010 Mar 11 21:12:33 |
| SAM_t06 target hidden Markov model | NP_510002_1.t06.w0.5.mod | 2010 Mar 11 21:12:38 |
| SAM_t06 multiple alignment---sequence logo (eps format) (pdf format) | NP_510002_1.t06.w0.5-logo | 2010 Mar 11 21:12:48 |
| SAM_t06 consensus sequence | NP_510002_1.t06.w0.5.maxp | 2010 Mar 11 21:12:49 |
| Script for highlighting in rasmol | conserved_t06 | 2010 Mar 11 21:12:49 |
| SAM_t04 multiple alignment in a2m format | NP_510002_1.t04.a2m.gz | 2010 Mar 11 21:56:02 |
| SAM_t04 multiple alignment in pretty html format | NP_510002_1.t04.pa.html | 2010 Mar 11 21:57:51 |
| SAM_t04 target hidden Markov model | NP_510002_1.t04.w0.5.mod | 2010 Mar 11 21:57:55 |
| SAM_t04 multiple alignment---sequence logo (eps format) (pdf format) | NP_510002_1.t04.w0.5-logo | 2010 Mar 11 21:58:06 |
| SAM_t04 consensus sequence | NP_510002_1.t04.w0.5.maxp | 2010 Mar 11 21:58:06 |
| Script for highlighting in rasmol | conserved_t04 | 2010 Mar 11 21:58:07 |
| SAM_t2k multiple alignment in a2m format | NP_510002_1.t2k.a2m.gz | 2010 Mar 11 22:18:44 |
| SAM_t2k multiple alignment in pretty html format | NP_510002_1.t2k.pa.html | 2010 Mar 11 22:20:36 |
| SAM_t2k target hidden Markov model | NP_510002_1.t2k.w0.5.mod | 2010 Mar 11 22:20:40 |
| SAM_t2k multiple alignment---sequence logo (eps format) (pdf format) | NP_510002_1.t2k.w0.5-logo | 2010 Mar 11 22:20:50 |
| SAM_t2k consensus sequence | NP_510002_1.t2k.w0.5.maxp | 2010 Mar 11 22:20:50 |
| Script for highlighting in rasmol | conserved_t2k | 2010 Mar 11 22:20:51 |
| Secondary Structure Prediction (Explanation of secondary-structure predictions) |
| str4 structure prediction (rdb format) (seq format) | NP_510002_1.t06.str4 | 2010 Mar 11 22:21:03 |
| Script for NP_510002_1.t06.str4 coloring in rasmol | NP_510002_1.t06.str4-color.rasmol | 2010 Mar 11 22:21:04 |
| str4---sequence logo (eps format) (pdf format) | NP_510002_1.t06.str4-logo | 2010 Mar 11 22:21:07 |
| str4 structure prediction (rdb format) (seq format) | NP_510002_1.t04.str4 | 2010 Mar 11 22:21:10 |
| Script for NP_510002_1.t04.str4 coloring in rasmol | NP_510002_1.t04.str4-color.rasmol | 2010 Mar 11 22:21:11 |
| str4---sequence logo (eps format) (pdf format) | NP_510002_1.t04.str4-logo | 2010 Mar 11 22:21:14 |
| str4 structure prediction (rdb format) (seq format) | NP_510002_1.t2k.str4 | 2010 Mar 11 22:21:16 |
| Script for NP_510002_1.t2k.str4 coloring in rasmol | NP_510002_1.t2k.str4-color.rasmol | 2010 Mar 11 22:21:17 |
| str4---sequence logo (eps format) (pdf format) | NP_510002_1.t2k.str4-logo | 2010 Mar 11 22:21:20 |
| str2 structure prediction (rdb format) (seq format) | NP_510002_1.t06.str2 | 2010 Mar 11 21:12:45 |
| Script for NP_510002_1.t06.str2 coloring in rasmol | NP_510002_1.t06.str2-color.rasmol | 2010 Mar 11 22:21:21 |
| str2---sequence logo (eps format) (pdf format) | NP_510002_1.t06.str2-logo | 2010 Mar 11 22:21:24 |
| str2 structure prediction (rdb format) (seq format) | NP_510002_1.t04.str2 | 2010 Mar 11 21:58:02 |
| Script for NP_510002_1.t04.str2 coloring in rasmol | NP_510002_1.t04.str2-color.rasmol | 2010 Mar 11 22:21:25 |
| str2---sequence logo (eps format) (pdf format) | NP_510002_1.t04.str2-logo | 2010 Mar 11 22:21:28 |
| str2 structure prediction (rdb format) (seq format) | NP_510002_1.t2k.str2 | 2010 Mar 11 22:20:46 |
| Script for NP_510002_1.t2k.str2 coloring in rasmol | NP_510002_1.t2k.str2-color.rasmol | 2010 Mar 11 22:21:28 |
| str2---sequence logo (eps format) (pdf format) | NP_510002_1.t2k.str2-logo | 2010 Mar 11 22:21:31 |
| alpha structure prediction (rdb format) (seq format) | NP_510002_1.t06.alpha | 2010 Mar 11 22:21:35 |
| Script for NP_510002_1.t06.alpha coloring in rasmol | NP_510002_1.t06.alpha-color.rasmol | 2010 Mar 11 22:21:35 |
| alpha---sequence logo (eps format) (pdf format) | NP_510002_1.t06.alpha-logo | 2010 Mar 11 22:21:38 |
| alpha structure prediction (rdb format) (seq format) | NP_510002_1.t04.alpha | 2010 Mar 11 22:21:42 |
| Script for NP_510002_1.t04.alpha coloring in rasmol | NP_510002_1.t04.alpha-color.rasmol | 2010 Mar 11 22:21:42 |
| alpha---sequence logo (eps format) (pdf format) | NP_510002_1.t04.alpha-logo | 2010 Mar 11 22:21:45 |
| alpha structure prediction (rdb format) (seq format) | NP_510002_1.t2k.alpha | 2010 Mar 11 22:21:49 |
| Script for NP_510002_1.t2k.alpha coloring in rasmol | NP_510002_1.t2k.alpha-color.rasmol | 2010 Mar 11 22:21:49 |
| alpha---sequence logo (eps format) (pdf format) | NP_510002_1.t2k.alpha-logo | 2010 Mar 11 22:21:52 |
| bys structure prediction (rdb format) (seq format) | NP_510002_1.t06.bys | 2010 Mar 11 22:21:56 |
| Script for NP_510002_1.t06.bys coloring in rasmol | NP_510002_1.t06.bys-color.rasmol | 2010 Mar 11 22:21:56 |
| bys---sequence logo (eps format) (pdf format) | NP_510002_1.t06.bys-logo | 2010 Mar 11 22:21:59 |
| bys structure prediction (rdb format) (seq format) | NP_510002_1.t04.bys | 2010 Mar 11 22:22:02 |
| Script for NP_510002_1.t04.bys coloring in rasmol | NP_510002_1.t04.bys-color.rasmol | 2010 Mar 11 22:22:02 |
| bys---sequence logo (eps format) (pdf format) | NP_510002_1.t04.bys-logo | 2010 Mar 11 22:22:05 |
| bys structure prediction (rdb format) (seq format) | NP_510002_1.t2k.bys | 2010 Mar 11 22:22:08 |
| Script for NP_510002_1.t2k.bys coloring in rasmol | NP_510002_1.t2k.bys-color.rasmol | 2010 Mar 11 22:22:08 |
| bys---sequence logo (eps format) (pdf format) | NP_510002_1.t2k.bys-logo | 2010 Mar 11 22:22:11 |
| pb structure prediction (rdb format) (seq format) | NP_510002_1.t06.pb | 2010 Mar 11 22:22:15 |
| Script for NP_510002_1.t06.pb coloring in rasmol | NP_510002_1.t06.pb-color.rasmol | 2010 Mar 11 22:22:15 |
| pb---sequence logo (eps format) (pdf format) | NP_510002_1.t06.pb-logo | 2010 Mar 11 22:22:18 |
| pb structure prediction (rdb format) (seq format) | NP_510002_1.t04.pb | 2010 Mar 11 22:22:22 |
| Script for NP_510002_1.t04.pb coloring in rasmol | NP_510002_1.t04.pb-color.rasmol | 2010 Mar 11 22:22:23 |
| pb---sequence logo (eps format) (pdf format) | NP_510002_1.t04.pb-logo | 2010 Mar 11 22:22:25 |
| pb structure prediction (rdb format) (seq format) | NP_510002_1.t2k.pb | 2010 Mar 11 22:22:30 |
| Script for NP_510002_1.t2k.pb coloring in rasmol | NP_510002_1.t2k.pb-color.rasmol | 2010 Mar 11 22:22:30 |
| pb---sequence logo (eps format) (pdf format) | NP_510002_1.t2k.pb-logo | 2010 Mar 11 22:22:33 |
| n_notor2 structure prediction (rdb format) (seq format) | NP_510002_1.t06.n_notor2 | 2010 Mar 11 22:22:37 |
| Script for NP_510002_1.t06.n_notor2 coloring in rasmol | NP_510002_1.t06.n_notor2-color.rasmol | 2010 Mar 11 22:22:38 |
| n_notor2---sequence logo (eps format) (pdf format) | NP_510002_1.t06.n_notor2-logo | 2010 Mar 11 22:22:41 |
| n_notor2 structure prediction (rdb format) (seq format) | NP_510002_1.t04.n_notor2 | 2010 Mar 11 22:22:45 |
| Script for NP_510002_1.t04.n_notor2 coloring in rasmol | NP_510002_1.t04.n_notor2-color.rasmol | 2010 Mar 11 22:22:46 |
| n_notor2---sequence logo (eps format) (pdf format) | NP_510002_1.t04.n_notor2-logo | 2010 Mar 11 22:22:49 |
| n_notor2 structure prediction (rdb format) (seq format) | NP_510002_1.t2k.n_notor2 | 2010 Mar 11 22:22:52 |
| Script for NP_510002_1.t2k.n_notor2 coloring in rasmol | NP_510002_1.t2k.n_notor2-color.rasmol | 2010 Mar 11 22:22:52 |
| n_notor2---sequence logo (eps format) (pdf format) | NP_510002_1.t2k.n_notor2-logo | 2010 Mar 11 22:22:55 |
| n_notor structure prediction (rdb format) (seq format) | NP_510002_1.t06.n_notor | 2010 Mar 11 22:22:59 |
| Script for NP_510002_1.t06.n_notor coloring in rasmol | NP_510002_1.t06.n_notor-color.rasmol | 2010 Mar 11 22:22:59 |
| n_notor---sequence logo (eps format) (pdf format) | NP_510002_1.t06.n_notor-logo | 2010 Mar 11 22:23:02 |
| n_notor structure prediction (rdb format) (seq format) | NP_510002_1.t04.n_notor | 2010 Mar 11 22:23:05 |
| Script for NP_510002_1.t04.n_notor coloring in rasmol | NP_510002_1.t04.n_notor-color.rasmol | 2010 Mar 11 22:23:06 |
| n_notor---sequence logo (eps format) (pdf format) | NP_510002_1.t04.n_notor-logo | 2010 Mar 11 22:23:08 |
| n_notor structure prediction (rdb format) (seq format) | NP_510002_1.t2k.n_notor | 2010 Mar 11 22:23:11 |
| Script for NP_510002_1.t2k.n_notor coloring in rasmol | NP_510002_1.t2k.n_notor-color.rasmol | 2010 Mar 11 22:23:13 |
| n_notor---sequence logo (eps format) (pdf format) | NP_510002_1.t2k.n_notor-logo | 2010 Mar 11 22:23:16 |
| n_sep structure prediction (rdb format) (seq format) | NP_510002_1.t06.n_sep | 2010 Mar 11 22:23:20 |
| Script for NP_510002_1.t06.n_sep coloring in rasmol | NP_510002_1.t06.n_sep-color.rasmol | 2010 Mar 11 22:23:20 |
| n_sep---sequence logo (eps format) (pdf format) | NP_510002_1.t06.n_sep-logo | 2010 Mar 11 22:23:24 |
| n_sep structure prediction (rdb format) (seq format) | NP_510002_1.t04.n_sep | 2010 Mar 11 22:23:28 |
| Script for NP_510002_1.t04.n_sep coloring in rasmol | NP_510002_1.t04.n_sep-color.rasmol | 2010 Mar 11 22:23:28 |
| n_sep---sequence logo (eps format) (pdf format) | NP_510002_1.t04.n_sep-logo | 2010 Mar 11 22:23:31 |
| n_sep structure prediction (rdb format) (seq format) | NP_510002_1.t2k.n_sep | 2010 Mar 11 22:23:35 |
| Script for NP_510002_1.t2k.n_sep coloring in rasmol | NP_510002_1.t2k.n_sep-color.rasmol | 2010 Mar 11 22:23:36 |
| n_sep---sequence logo (eps format) (pdf format) | NP_510002_1.t2k.n_sep-logo | 2010 Mar 11 22:23:38 |
| o_notor2 structure prediction (rdb format) (seq format) | NP_510002_1.t06.o_notor2 | 2010 Mar 11 22:23:42 |
| Script for NP_510002_1.t06.o_notor2 coloring in rasmol | NP_510002_1.t06.o_notor2-color.rasmol | 2010 Mar 11 22:23:42 |
| o_notor2---sequence logo (eps format) (pdf format) | NP_510002_1.t06.o_notor2-logo | 2010 Mar 11 22:23:45 |
| o_notor2 structure prediction (rdb format) (seq format) | NP_510002_1.t04.o_notor2 | 2010 Mar 11 22:23:48 |
| Script for NP_510002_1.t04.o_notor2 coloring in rasmol | NP_510002_1.t04.o_notor2-color.rasmol | 2010 Mar 11 22:23:48 |
| o_notor2---sequence logo (eps format) (pdf format) | NP_510002_1.t04.o_notor2-logo | 2010 Mar 11 22:23:52 |
| o_notor2 structure prediction (rdb format) (seq format) | NP_510002_1.t2k.o_notor2 | 2010 Mar 11 22:23:57 |
| Script for NP_510002_1.t2k.o_notor2 coloring in rasmol | NP_510002_1.t2k.o_notor2-color.rasmol | 2010 Mar 11 22:23:57 |
| o_notor2---sequence logo (eps format) (pdf format) | NP_510002_1.t2k.o_notor2-logo | 2010 Mar 11 22:24:00 |
| o_notor structure prediction (rdb format) (seq format) | NP_510002_1.t06.o_notor | 2010 Mar 11 22:24:04 |
| Script for NP_510002_1.t06.o_notor coloring in rasmol | NP_510002_1.t06.o_notor-color.rasmol | 2010 Mar 11 22:24:04 |
| o_notor---sequence logo (eps format) (pdf format) | NP_510002_1.t06.o_notor-logo | 2010 Mar 11 22:24:07 |
| o_notor structure prediction (rdb format) (seq format) | NP_510002_1.t04.o_notor | 2010 Mar 11 22:24:10 |
| Script for NP_510002_1.t04.o_notor coloring in rasmol | NP_510002_1.t04.o_notor-color.rasmol | 2010 Mar 11 22:24:10 |
| o_notor---sequence logo (eps format) (pdf format) | NP_510002_1.t04.o_notor-logo | 2010 Mar 11 22:24:13 |
| o_notor structure prediction (rdb format) (seq format) | NP_510002_1.t2k.o_notor | 2010 Mar 11 22:24:15 |
| Script for NP_510002_1.t2k.o_notor coloring in rasmol | NP_510002_1.t2k.o_notor-color.rasmol | 2010 Mar 11 22:24:16 |
| o_notor---sequence logo (eps format) (pdf format) | NP_510002_1.t2k.o_notor-logo | 2010 Mar 11 22:24:19 |
| o_sep structure prediction (rdb format) (seq format) | NP_510002_1.t06.o_sep | 2010 Mar 11 22:24:24 |
| Script for NP_510002_1.t06.o_sep coloring in rasmol | NP_510002_1.t06.o_sep-color.rasmol | 2010 Mar 11 22:24:25 |
| o_sep---sequence logo (eps format) (pdf format) | NP_510002_1.t06.o_sep-logo | 2010 Mar 11 22:24:29 |
| o_sep structure prediction (rdb format) (seq format) | NP_510002_1.t04.o_sep | 2010 Mar 11 22:24:32 |
| Script for NP_510002_1.t04.o_sep coloring in rasmol | NP_510002_1.t04.o_sep-color.rasmol | 2010 Mar 11 22:24:32 |
| o_sep---sequence logo (eps format) (pdf format) | NP_510002_1.t04.o_sep-logo | 2010 Mar 11 22:24:36 |
| o_sep structure prediction (rdb format) (seq format) | NP_510002_1.t2k.o_sep | 2010 Mar 11 22:24:39 |
| Script for NP_510002_1.t2k.o_sep coloring in rasmol | NP_510002_1.t2k.o_sep-color.rasmol | 2010 Mar 11 22:24:39 |
| o_sep---sequence logo (eps format) (pdf format) | NP_510002_1.t2k.o_sep-logo | 2010 Mar 11 22:24:42 |
| dssp-ehl2 structure prediction (rdb format) (seq format) | NP_510002_1.t06.dssp-ehl2 | 2010 Mar 11 22:24:44 |
| Script for NP_510002_1.t06.dssp-ehl2 coloring in rasmol | NP_510002_1.t06.dssp-ehl2-color.rasmol | 2010 Mar 11 22:24:44 |
| dssp-ehl2---sequence logo (eps format) (pdf format) | NP_510002_1.t06.dssp-ehl2-logo | 2010 Mar 11 22:24:47 |
| dssp-ehl2 structure prediction (rdb format) (seq format) | NP_510002_1.t04.dssp-ehl2 | 2010 Mar 11 22:24:48 |
| Script for NP_510002_1.t04.dssp-ehl2 coloring in rasmol | NP_510002_1.t04.dssp-ehl2-color.rasmol | 2010 Mar 11 22:24:49 |
| dssp-ehl2---sequence logo (eps format) (pdf format) | NP_510002_1.t04.dssp-ehl2-logo | 2010 Mar 11 22:24:51 |
| dssp-ehl2 structure prediction (rdb format) (seq format) | NP_510002_1.t2k.dssp-ehl2 | 2010 Mar 11 22:24:53 |
| Script for NP_510002_1.t2k.dssp-ehl2 coloring in rasmol | NP_510002_1.t2k.dssp-ehl2-color.rasmol | 2010 Mar 11 22:24:53 |
| dssp-ehl2---sequence logo (eps format) (pdf format) | NP_510002_1.t2k.dssp-ehl2-logo | 2010 Mar 11 22:24:55 |
| dssp-ehl2 structure prediction (rdb format) (seq format) | NP_510002_1.dssp-ehl2 | 2010 Mar 11 22:24:58 |
| Script for NP_510002_1.dssp-ehl2 coloring in rasmol | NP_510002_1.dssp-ehl2-color.rasmol | 2010 Mar 11 22:24:58 |
| dssp-ehl2---sequence logo (eps format) (pdf format) | NP_510002_1.dssp-ehl2-logo | 2010 Mar 11 22:25:00 |
| CB_burial_14_7 structure prediction (rdb format) (seq format) | NP_510002_1.t06.CB_burial_14_7 | 2010 Mar 11 22:25:05 |
| Script for NP_510002_1.t06.CB_burial_14_7 coloring in rasmol | NP_510002_1.t06.CB_burial_14_7-color.rasmol | 2010 Mar 11 22:25:06 |
| CB_burial_14_7---sequence logo (eps format) (pdf format) | NP_510002_1.t06.CB_burial_14_7-logo | 2010 Mar 11 22:25:09 |
| CB_burial_14_7 structure prediction (rdb format) (seq format) | NP_510002_1.t04.CB_burial_14_7 | 2010 Mar 11 22:25:12 |
| Script for NP_510002_1.t04.CB_burial_14_7 coloring in rasmol | NP_510002_1.t04.CB_burial_14_7-color.rasmol | 2010 Mar 11 22:25:12 |
| CB_burial_14_7---sequence logo (eps format) (pdf format) | NP_510002_1.t04.CB_burial_14_7-logo | 2010 Mar 11 22:25:15 |
| CB_burial_14_7 structure prediction (rdb format) (seq format) | NP_510002_1.t2k.CB_burial_14_7 | 2010 Mar 11 22:25:18 |
| Script for NP_510002_1.t2k.CB_burial_14_7 coloring in rasmol | NP_510002_1.t2k.CB_burial_14_7-color.rasmol | 2010 Mar 11 22:25:18 |
| CB_burial_14_7---sequence logo (eps format) (pdf format) | NP_510002_1.t2k.CB_burial_14_7-logo | 2010 Mar 11 22:25:21 |
| near-backbone-11 structure prediction (rdb format) (seq format) | NP_510002_1.t06.near-backbone-11 | 2010 Mar 11 22:25:25 |
| Script for NP_510002_1.t06.near-backbone-11 coloring in rasmol | NP_510002_1.t06.near-backbone-11-color.rasmol | 2010 Mar 11 22:25:25 |
| near-backbone-11---sequence logo (eps format) (pdf format) | NP_510002_1.t06.near-backbone-11-logo | 2010 Mar 11 22:25:28 |
| near-backbone-11 structure prediction (rdb format) (seq format) | NP_510002_1.t04.near-backbone-11 | 2010 Mar 11 22:25:31 |
| Script for NP_510002_1.t04.near-backbone-11 coloring in rasmol | NP_510002_1.t04.near-backbone-11-color.rasmol | 2010 Mar 11 22:25:31 |
| near-backbone-11---sequence logo (eps format) (pdf format) | NP_510002_1.t04.near-backbone-11-logo | 2010 Mar 11 22:25:34 |
| near-backbone-11 structure prediction (rdb format) (seq format) | NP_510002_1.t2k.near-backbone-11 | 2010 Mar 11 22:25:37 |
| Script for NP_510002_1.t2k.near-backbone-11 coloring in rasmol | NP_510002_1.t2k.near-backbone-11-color.rasmol | 2010 Mar 11 22:25:38 |
| near-backbone-11---sequence logo (eps format) (pdf format) | NP_510002_1.t2k.near-backbone-11-logo | 2010 Mar 11 22:25:42 |
| CB8-sep9 structure prediction (rdb format) (seq format) | NP_510002_1.t06.CB8-sep9 | 2010 Mar 11 22:25:47 |
| Script for NP_510002_1.t06.CB8-sep9 coloring in rasmol | NP_510002_1.t06.CB8-sep9-color.rasmol | 2010 Mar 11 22:25:47 |
| CB8-sep9---sequence logo (eps format) (pdf format) | NP_510002_1.t06.CB8-sep9-logo | 2010 Mar 11 22:25:51 |
| CB8-sep9 structure prediction (rdb format) (seq format) | NP_510002_1.t04.CB8-sep9 | 2010 Mar 11 22:25:54 |
| Script for NP_510002_1.t04.CB8-sep9 coloring in rasmol | NP_510002_1.t04.CB8-sep9-color.rasmol | 2010 Mar 11 22:25:55 |
| CB8-sep9---sequence logo (eps format) (pdf format) | NP_510002_1.t04.CB8-sep9-logo | 2010 Mar 11 22:25:58 |
| CB8-sep9 structure prediction (rdb format) (seq format) | NP_510002_1.t2k.CB8-sep9 | 2010 Mar 11 22:26:02 |
| Script for NP_510002_1.t2k.CB8-sep9 coloring in rasmol | NP_510002_1.t2k.CB8-sep9-color.rasmol | 2010 Mar 11 22:26:02 |
| CB8-sep9---sequence logo (eps format) (pdf format) | NP_510002_1.t2k.CB8-sep9-logo | 2010 Mar 11 22:26:05 |
| Target model scores |
| amino acid single-track target model scores of PDB | NP_510002_1.t06-w0.5-scores.html | 2010 Mar 11 22:36:31 |
| amino acid single-track target model scores of PDB | NP_510002_1.t04-w0.5-scores.html | 2010 Mar 11 22:46:22 |
| amino acid single-track target model scores of PDB | NP_510002_1.t2k-w0.5-scores.html | 2010 Mar 11 22:56:06 |
| amino acid/w0.5-1-str4-0.1 multi-track target model scores | NP_510002_1.t06-w0.5-1-str4-0.1-scores.html | 2010 Mar 11 23:01:39 |
| amino acid/w0.5-1-str2-0.1 multi-track target model scores | NP_510002_1.t06-w0.5-1-str2-0.1-scores.html | 2010 Mar 11 23:07:15 |
| amino acid/w0.5-1-alpha-0.1 multi-track target model scores | NP_510002_1.t06-w0.5-1-alpha-0.1-scores.html | 2010 Mar 11 23:12:46 |
| amino acid/w0.5-1-bys-0.1 multi-track target model scores | NP_510002_1.t06-w0.5-1-bys-0.1-scores.html | 2010 Mar 11 23:18:18 |
| amino acid/w0.5-1-pb-0.1 multi-track target model scores | NP_510002_1.t06-w0.5-1-pb-0.1-scores.html | 2010 Mar 11 23:23:54 |
| amino acid/w0.5-1-n_notor2-0.1 multi-track target model scores | NP_510002_1.t06-w0.5-1-n_notor2-0.1-scores.html | 2010 Mar 11 23:29:24 |
| amino acid/w0.5-1-n_notor-0.1 multi-track target model scores | NP_510002_1.t06-w0.5-1-n_notor-0.1-scores.html | 2010 Mar 11 23:34:56 |
| amino acid/w0.5-1-n_sep-0.1 multi-track target model scores | NP_510002_1.t06-w0.5-1-n_sep-0.1-scores.html | 2010 Mar 11 23:40:28 |
| amino acid/w0.5-1-o_notor2-0.1 multi-track target model scores | NP_510002_1.t06-w0.5-1-o_notor2-0.1-scores.html | 2010 Mar 11 23:46:02 |
| amino acid/w0.5-1-o_notor-0.1 multi-track target model scores | NP_510002_1.t06-w0.5-1-o_notor-0.1-scores.html | 2010 Mar 11 23:51:34 |
| amino acid/w0.5-1-o_sep-0.1 multi-track target model scores | NP_510002_1.t06-w0.5-1-o_sep-0.1-scores.html | 2010 Mar 11 23:57:10 |
| amino acid/w0.5-1-dssp-ehl2-0.1 multi-track target model scores | NP_510002_1.t06-w0.5-1-dssp-ehl2-0.1-scores.html | 2010 Mar 12 0:02:42 |
| amino acid/w0.5-1-CB_burial_14_7-0.3 multi-track target model scores | NP_510002_1.t06-w0.5-1-CB_burial_14_7-0.3-scores.html | 2010 Mar 12 0:08:16 |
| amino acid/w0.5-1-near-backbone-11-0.3 multi-track target model scores | NP_510002_1.t06-w0.5-1-near-backbone-11-0.3-scores.html | 2010 Mar 12 0:13:51 |
| amino acid/w0.5-1-CB8-sep9-0.3 multi-track target model scores | NP_510002_1.t06-w0.5-1-CB8-sep9-0.3-scores.html | 2010 Mar 12 0:19:24 |
| amino acid/80-60-80-str2+near-backbone-11 multi-track target model scores | NP_510002_1.t06-80-60-80-str2+near-backbone-11-scores.html | 2010 Mar 12 0:25:31 |
| amino acid/w0.5-1-str4-0.1 multi-track target model scores | NP_510002_1.t04-w0.5-1-str4-0.1-scores.html | 2010 Mar 12 0:31:33 |
| amino acid/w0.5-1-str2-0.1 multi-track target model scores | NP_510002_1.t04-w0.5-1-str2-0.1-scores.html | 2010 Mar 12 0:37:35 |
| amino acid/w0.5-1-alpha-0.1 multi-track target model scores | NP_510002_1.t04-w0.5-1-alpha-0.1-scores.html | 2010 Mar 12 0:43:40 |
| amino acid/w0.5-1-bys-0.1 multi-track target model scores | NP_510002_1.t04-w0.5-1-bys-0.1-scores.html | 2010 Mar 12 0:49:45 |
| amino acid/w0.5-1-pb-0.1 multi-track target model scores | NP_510002_1.t04-w0.5-1-pb-0.1-scores.html | 2010 Mar 12 0:55:47 |
| amino acid/w0.5-1-n_notor2-0.1 multi-track target model scores | NP_510002_1.t04-w0.5-1-n_notor2-0.1-scores.html | 2010 Mar 12 1:01:50 |
| amino acid/w0.5-1-n_notor-0.1 multi-track target model scores | NP_510002_1.t04-w0.5-1-n_notor-0.1-scores.html | 2010 Mar 12 1:07:49 |
| amino acid/w0.5-1-n_sep-0.1 multi-track target model scores | NP_510002_1.t04-w0.5-1-n_sep-0.1-scores.html | 2010 Mar 12 1:13:53 |
| amino acid/w0.5-1-o_notor2-0.1 multi-track target model scores | NP_510002_1.t04-w0.5-1-o_notor2-0.1-scores.html | 2010 Mar 12 1:19:56 |
| amino acid/w0.5-1-o_notor-0.1 multi-track target model scores | NP_510002_1.t04-w0.5-1-o_notor-0.1-scores.html | 2010 Mar 12 1:25:56 |
| amino acid/w0.5-1-o_sep-0.1 multi-track target model scores | NP_510002_1.t04-w0.5-1-o_sep-0.1-scores.html | 2010 Mar 12 1:32:01 |
| amino acid/w0.5-1-dssp-ehl2-0.1 multi-track target model scores | NP_510002_1.t04-w0.5-1-dssp-ehl2-0.1-scores.html | 2010 Mar 12 1:38:02 |
| amino acid/w0.5-1-CB_burial_14_7-0.3 multi-track target model scores | NP_510002_1.t04-w0.5-1-CB_burial_14_7-0.3-scores.html | 2010 Mar 12 1:44:11 |
| amino acid/w0.5-1-near-backbone-11-0.3 multi-track target model scores | NP_510002_1.t04-w0.5-1-near-backbone-11-0.3-scores.html | 2010 Mar 12 1:50:15 |
| amino acid/w0.5-1-CB8-sep9-0.3 multi-track target model scores | NP_510002_1.t04-w0.5-1-CB8-sep9-0.3-scores.html | 2010 Mar 12 1:56:15 |
| amino acid/80-60-80-str2+near-backbone-11 multi-track target model scores | NP_510002_1.t04-80-60-80-str2+near-backbone-11-scores.html | 2010 Mar 12 2:02:54 |
| amino acid/w0.5-1-str4-0.1 multi-track target model scores | NP_510002_1.t2k-w0.5-1-str4-0.1-scores.html | 2010 Mar 12 2:09:30 |
| amino acid/w0.5-1-str2-0.1 multi-track target model scores | NP_510002_1.t2k-w0.5-1-str2-0.1-scores.html | 2010 Mar 12 2:16:07 |
| amino acid/w0.5-1-alpha-0.1 multi-track target model scores | NP_510002_1.t2k-w0.5-1-alpha-0.1-scores.html | 2010 Mar 12 2:22:40 |
| amino acid/w0.5-1-bys-0.1 multi-track target model scores | NP_510002_1.t2k-w0.5-1-bys-0.1-scores.html | 2010 Mar 12 2:29:20 |
| amino acid/w0.5-1-pb-0.1 multi-track target model scores | NP_510002_1.t2k-w0.5-1-pb-0.1-scores.html | 2010 Mar 12 2:35:56 |
| amino acid/w0.5-1-n_notor2-0.1 multi-track target model scores | NP_510002_1.t2k-w0.5-1-n_notor2-0.1-scores.html | 2010 Mar 12 2:42:31 |
| amino acid/w0.5-1-n_notor-0.1 multi-track target model scores | NP_510002_1.t2k-w0.5-1-n_notor-0.1-scores.html | 2010 Mar 12 2:49:06 |
| amino acid/w0.5-1-n_sep-0.1 multi-track target model scores | NP_510002_1.t2k-w0.5-1-n_sep-0.1-scores.html | 2010 Mar 12 2:55:44 |
| amino acid/w0.5-1-o_notor2-0.1 multi-track target model scores | NP_510002_1.t2k-w0.5-1-o_notor2-0.1-scores.html | 2010 Mar 12 3:02:21 |
| amino acid/w0.5-1-o_notor-0.1 multi-track target model scores | NP_510002_1.t2k-w0.5-1-o_notor-0.1-scores.html | 2010 Mar 12 3:08:57 |
| amino acid/w0.5-1-o_sep-0.1 multi-track target model scores | NP_510002_1.t2k-w0.5-1-o_sep-0.1-scores.html | 2010 Mar 12 3:15:36 |
| amino acid/w0.5-1-dssp-ehl2-0.1 multi-track target model scores | NP_510002_1.t2k-w0.5-1-dssp-ehl2-0.1-scores.html | 2010 Mar 12 3:22:10 |
| amino acid/w0.5-1-CB_burial_14_7-0.3 multi-track target model scores | NP_510002_1.t2k-w0.5-1-CB_burial_14_7-0.3-scores.html | 2010 Mar 12 3:28:49 |
| amino acid/w0.5-1-near-backbone-11-0.3 multi-track target model scores | NP_510002_1.t2k-w0.5-1-near-backbone-11-0.3-scores.html | 2010 Mar 12 3:35:31 |
| amino acid/w0.5-1-CB8-sep9-0.3 multi-track target model scores | NP_510002_1.t2k-w0.5-1-CB8-sep9-0.3-scores.html | 2010 Mar 12 3:42:09 |
| amino acid/80-60-80-str2+near-backbone-11 multi-track target model scores | NP_510002_1.t2k-80-60-80-str2+near-backbone-11-scores.html | 2010 Mar 12 3:49:19 |
| Template model scores |
| Annotated t06 template model scores | NP_510002_1.t06-template-lib-scores.html | 2010 Mar 12 4:52:52 |
| Annotated t04 template model scores | NP_510002_1.t04-template-lib-scores.html | 2010 Mar 12 5:59:43 |
| Annotated t2k template model scores | NP_510002_1.t2k-template-lib-scores.html | 2010 Mar 12 6:42:27 |
| Top Hits |
| Best scoring hits from t06 HMMs | NP_510002_1.t06.best-scores.html | 2010 Mar 12 6:52:41 |
| Best scoring hits from t04 HMMs | NP_510002_1.t04.best-scores.html | 2010 Mar 12 7:02:32 |
| Best scoring hits from t2k HMMs | NP_510002_1.t2k.best-scores.html | 2010 Mar 12 7:12:20 |
| Best scoring hits from combining t06 t04 t2k | NP_510002_1.best-scores-all.html | 2010 Mar 12 7:12:21 |
| Alignments for Top Hits |
| Top alignments from t06 hits | NP_510002_1.t06.top_reported_alignments.html | 2010 Mar 12 8:02:44 |
| Top alignments from t04 hits | NP_510002_1.t04.top_reported_alignments.html | 2010 Mar 12 8:02:45 |
| Top alignments from t2k hits | NP_510002_1.t2k.top_reported_alignments.html | 2010 Mar 12 8:02:46 |
| Top alignments from combined hits | NP_510002_1.top_reported_alignments.html | 2010 Mar 12 8:02:48 |
| multiple alignment of templates (a2m.gz format) (pa format) | all-align | 2010 Mar 12 8:03:29 |
| Undertaker (3d) files |
| Undertaker input for top t06 alignments | NP_510002_1.t06.undertaker-align.under | 2010 Mar 12 8:05:57 |
| Undertaker input for top t04 alignments | NP_510002_1.t04.undertaker-align.under | 2010 Mar 12 8:05:58 |
| Undertaker input for top t2k alignments | NP_510002_1.t2k.undertaker-align.under | 2010 Mar 12 8:05:59 |
| Undertaker input for top combined alignments | NP_510002_1.undertaker-align.under | 2010 Mar 12 8:06:00 |
| PDB file with model(s) for top alignments | NP_510002_1.undertaker-align.pdb | 2010 Mar 12 8:08:24 |
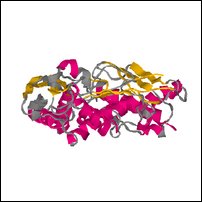
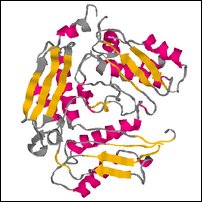
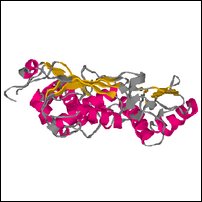
Images of an (incomplete) model created by sidechain replacement on the backbone of the highest scoring template.
Coloring is defined by ehl2, to show predicted secondary structure (red for helix, yellow for strand, blue for turn, grey for other).
| Front | Top | Side |
 |
 |
 |
| 500x500 | 500x500 | 500x500 |
| Fragment list for undertaker (from fragfinder) | NP_510002_1.t06.many.frag.gz | 2010 Mar 12 8:10:13 |
| Fragment list for undertaker (from fragfinder) | NP_510002_1.t04.many.frag.gz | 2010 Mar 12 8:11:52 |
| Fragment list for undertaker (from fragfinder) | NP_510002_1.t2k.many.frag.gz | 2010 Mar 12 8:13:31 |
| Contact predictions, based on |
| contact predictions (rr format) (rr.nn1000.constraints format) (rr.rasmol format) | NP_510002_1.647_47 | 2010 Mar 12 8:20:00 |
| contact predictions (rr format) (rr.nn1000.constraints format) (rr.rasmol format) | NP_510002_1.730_47 | 2010 Mar 12 8:20:16 |
| contact predictions (rr format) (rr.nn1000.constraints format) (rr.rasmol format) | NP_510002_1.648_17.730_47 | 2010 Mar 12 8:20:54 |
| Distance constraints extracted from alignments} |
| Constraints from alignment | align.constraints | 2010 Mar 12 8:24:37 |
| Rejected constraints | rejected_bonus.constraints | 2010 Mar 12 8:24:37 |
| Noncontact constraints | noncontact.constraints | 2010 Mar 12 8:24:37 |
| First attempted model using undertaker | decoys/NP_510002_1.try1-opt3.pdb | 2010 Mar 12 11:35:34 |
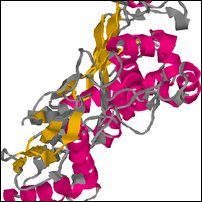
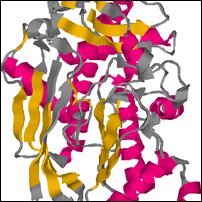
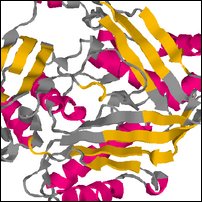
Images of complete model in decoys/NP_510002_1.try1-opt3.pdb
Coloring is defined by the dssp rasmol script, to show predicted secondary structure (red for helix, yellow for strand, blue for turn, grey for other).
| Front | Top | Side |
 |
 |
 |
| 500x500 | 500x500 | 500x500 |
| Gzipped tarball of all the files | NP_510002_1.tgz | 2010 Mar 12 11:42:00 |