Objectives:
- Use Entrez in multiple ways to find DNA or protein sequences of interest
- Appreciate file format differences and rudiments of file conversion
- Learn to import Genbank files into VectorNTI
- Learn to conduct simple manipulations in VectorNTI corresponding to fundamental molecular biology questions.
- Review basic aspects of biology that will come up later in the course.
Remember: e-mail your lab to Yeshi (tgyeshi@uky.edu) with a subject line "BIO520 Lab 2". Use Rich Text Format (.rtf) or MS Word format (.doc) and name the document like so: LundJ_lab2.rtf. Do not use the 'new' MS Word format that gives files a .docx extension!
The best thing to do is to start work on lab 2 and look at the reference material as needed. For the biological review questions, the lab 1 biology reference pages are very handy.
- 1. Please use Entrez (the Entrez Help is very helpful!) to answer the following questions. Find the nucleotide sequence of the human hemoglobin alpha 1 gene.:
a. Search for this gene in the Nucleotide section of Genbank (http://www.ncbi.nlm.nih.gov/entrez). Doing a search for "hemoglobin" how many Nucleotide sequences are returned (ignore EST and GSS sequences for this question)? ANSWER=number of sequences
Looking at these sequences, you can see that most are not hemoglobins! Why is this?b. Now search in Genbank using hemoglobin as a "Title" word (Search Field Help). How many sequences are returned? ANSWER=number of sequences
c. Now constrain your search further to find the RefSeq entry for human hemoglobin alpha 1. ANSWER=accession number (Keep this search page active, you'll use it in question 2).
d. Type here the search string that finds PROTEIN sequences from the organism Saccharomyces cerevisiae whose entries contain the phrase "transcription factor". This is a Boolean type query. There are several ways to execute it and the number of entries found can vary.
e. How many entries do you find (in the "All:" tab)? ANSWER=number Keep this search active for question 1f.
f. Now, do this same search in the Structure database-how many database entries are there for yeast proteins whose entries are found with a search for "transcription factor"? ANSWER=number? Several proteins have multiple structures in the database so the number of yeast TFs with structures is lower. Entrez can allow you to go from one type of information to another very quickly.
- 2. Follow the link to the Entrez Gene entry for human alpha 1 hemoglobin.
a. Using the information on this page, give a description of the function of hemoglobin. ANSWER=a couple of words.
b. What conserved domain does this protein contain?
c. Is this domain present in microorganisms?
d. Give the OMIM entry number for a human genetic disease associated with this gene (their is more than one possible answer). ANSWER=number.
e. Follow the link to the MapViewer (Genome links also take you to this). On what human chromsome is hemoglobin alpha 1 found?
f. Find the genes which are next to hemoglobin alpha 1 on the chromosome (active genes, not pseudogenes). Give the two closest upstream and the two closest downstream genes.
Upstream refers to the region 5' of the gene. - 3. Download the human hemoglobin alpha 1 gene sequence entry from Genbank in two different formats: Genbank and FASTA. Save them with different names (include the accession number in the name). Open each file in a text editor (Notepad should work, or use MS Word if you prefer). Scroll through the files.
a. What is the approved symbol for this gene? ANSWER=gene symbol
Note which format has more biological information in it. Now, open each file in the Artemis program--be sure NOT to save the files as Word documents! PASTE here the following:
b. The graphics pane from the FASTA format file
c. The graphics pane from the Genbank file.
- 4. Use Artemis to search for open reading frames (ORFs) in this sequence.
a. In what frame is hemoglobin's ORF? ANSWER=(+1, +2, +3, -1, -2, or -3)
b. Translate the hemoglobin ORF. What stop codon is used?
- 5. List the amino acids with ACIDIC side chains (these have a negative charge at neutral pH) in an alphabetic string in one-letter code (eg. ACDE) You SHOULD know all 20 amino acids and their properties. The 1-letter code is easiest to use.
- 6. For a typical prokaryotic operon containing two cistrons shown below, list the indicated elements in the 5' to 3' order. For two elements that overlap indicate the one the starts most 5' first. The elements are indicated on the drawing (click on the drawing for a full-scale version). Each term is used only once, although each gene could have more than one of some of these features. You MUST know the structure of a typical gene!!
List the indicated elements as a string in 5' to 3' order.
A. Ribosome Binding Site-cistron A 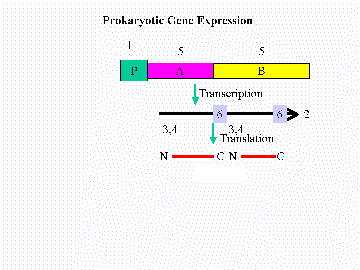
B. Promoter C. cistronA (ORF) D. cistron B E. Initiation codon-A F. Stop codon-B - 7. Estimate the following basic "biological ballpark"
quantities (answer as a string): HELP
-
A. Genome size of the human genome (approximate).
-
B. Genome size of E. coli genome (approximate).
-
C. Number of amino acids in a "typical" protein.
-
D. Molecular weight of an "average" amino acid.
-
E. Size, in kbp, of a "typical" bacterial cistron.
-
F. Size, in kbp, of a "typical" human gene.
-
G. Number of genes in the E.coli genome.
-
H. Number of genes in the human genome.
These figures help you navigate the biological world-they give you an order of magnitude sense of what is a reasonable answer when confronting a new problem.
-
- 8. You sequence the genes encoding
Glyceraldehyde-3-phosphate dehydrogenase from a fish, a rodent, a
primate, and from yeast.
-
A. Which pair do you think would be most similar in amino acid sequence? Answer is a pair, like yeast-fish.
-
B. Which of these genes do you think would be most different from all of the others? Answer is a single gene, like fish.
-
- 9. What experiments would you use to find the answer to the following biological questions? Answer=simple description, like Western blot:
-
A. You want to know the subunit molecular weight of a purified protein.
-
B. You want to find out what splice forms of a gene are expressed in heart, liver, muscle, and kidney.
-
C. You want a copy of the DNA sequence of a mutant version of a short gene whose wild type DNA sequence you already know.
-